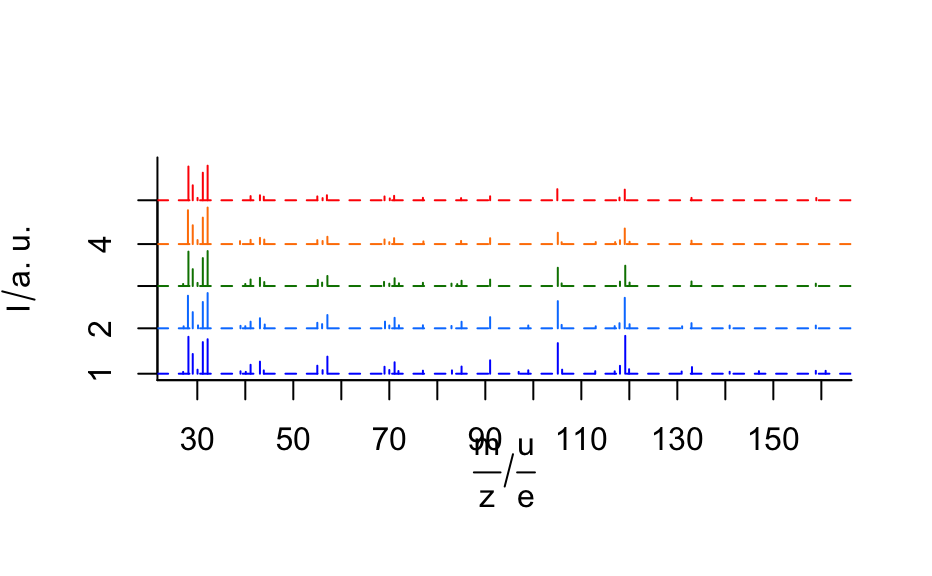
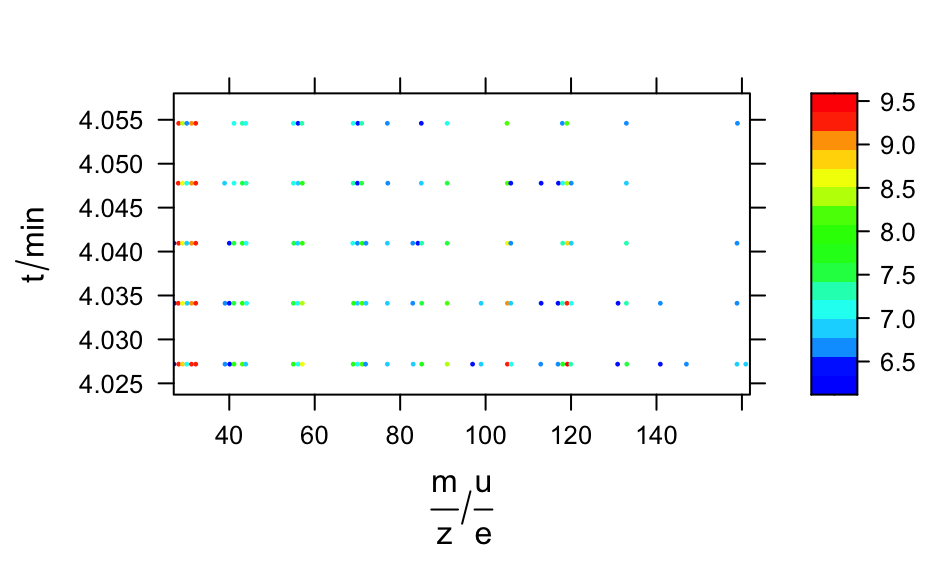
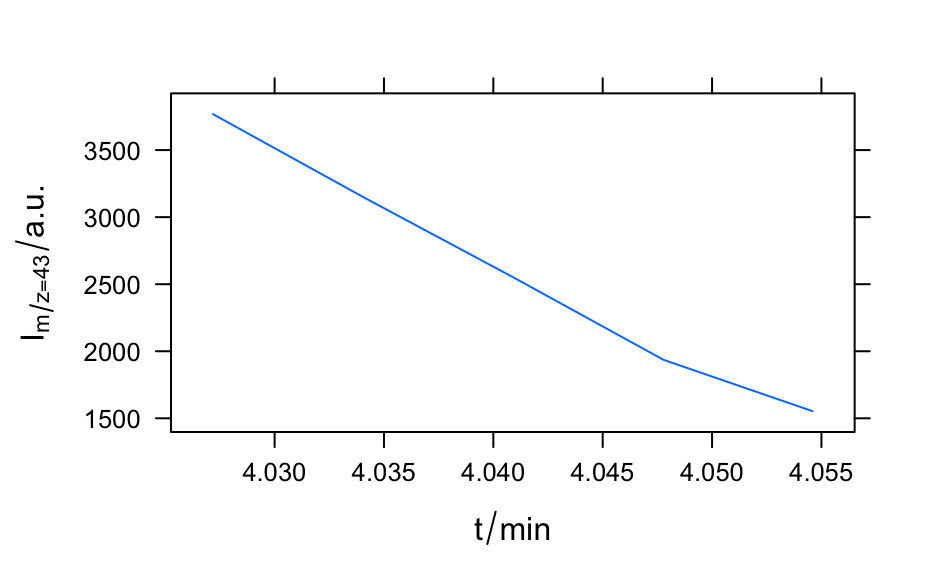
A time series of mass spectra in a list of hyperSpec objects.
barbiturates
Format
The data sets consists of a list of 5 hyperSpec objects with a single spectrum each. They are the first five spectra of the BARBITUATES.SPC example data from Thermo Galactic's spc file format specification.
References
The raw data is available with package hySpc.read.spc.
Author
C. Beleites and Thermo Galactic
Examples
barbiturates#> [[1]] #> hyperSpec object #> 1 spectra #> 4 data columns #> 37 data points / spectrum #> wavelength: frac(m, z)/frac(u, e) [numeric] 160.90 158.85 ... 27.05 #> data: (1 rows x 4 columns) #> 1. z: t/min [numeric] 4.027184 #> 2. z.end: t/min [numeric] 4.027184 #> 3. spc: I/"a. u." [matrix, array37] 902 896 ... 562 #> 4. filename: filename [character] /home/cb/R/x86_64-pc-linux-gnu-library/3.6/hyperSpec/extdata/BARBITUATES.SPC #> #> [[2]] #> hyperSpec object #> 1 spectra #> 4 data columns #> 34 data points / spectrum #> wavelength: frac(m, z)/frac(u, e) [numeric] 158.85 140.90 ... 27.15 #> data: (1 rows x 4 columns) #> 1. z: t/min [numeric] 4.034117 #> 2. z.end: t/min [numeric] 4.034117 #> 3. spc: I/"a. u." [matrix, array34] 857 864 ... 618 #> 4. filename: filename [character] /home/cb/R/x86_64-pc-linux-gnu-library/3.6/hyperSpec/extdata/BARBITUATES.SPC #> #> [[3]] #> hyperSpec object #> 1 spectra #> 4 data columns #> 29 data points / spectrum #> wavelength: frac(m, z)/frac(u, e) [numeric] 158.85 132.95 ... 27.05 #> data: (1 rows x 4 columns) #> 1. z: t/min [numeric] 4.04095 #> 2. z.end: t/min [numeric] 4.04095 #> 3. spc: I/"a. u." [matrix, array29] 735 1412 ... 638 #> 4. filename: filename [character] /home/cb/R/x86_64-pc-linux-gnu-library/3.6/hyperSpec/extdata/BARBITUATES.SPC #> #> [[4]] #> hyperSpec object #> 1 spectra #> 4 data columns #> 26 data points / spectrum #> wavelength: frac(m, z)/frac(u, e) [numeric] 132.95 120.05 ... 28.05 #> data: (1 rows x 4 columns) #> 1. z: t/min [numeric] 4.047783 #> 2. z.end: t/min [numeric] 4.047783 #> 3. spc: I/"a. u." [matrix, array26] 1066 718 ... 10548 #> 4. filename: filename [character] /home/cb/R/x86_64-pc-linux-gnu-library/3.6/hyperSpec/extdata/BARBITUATES.SPC #> #> [[5]] #> hyperSpec object #> 1 spectra #> 4 data columns #> 22 data points / spectrum #> wavelength: frac(m, z)/frac(u, e) [numeric] 158.95 132.95 ... 28.15 #> data: (1 rows x 4 columns) #> 1. z: t/min [numeric] 4.0546 #> 2. z.end: t/min [numeric] 4.0546 #> 3. spc: I/"a. u." [matrix, array22] 704 738 ... 10519 #> 4. filename: filename [character] /home/cb/R/x86_64-pc-linux-gnu-library/3.6/hyperSpec/extdata/BARBITUATES.SPC #>length(barbiturates)#> [1] 5barb <- collapse(barbiturates, collapse.equal = FALSE) barb <- orderwl(barb) plot(barb, lines.args = list(type = "h"), col = matlab.dark.palette(5), stacked = TRUE, stacked.args = list(add.factor = .2) )if (require(latticeExtra)) { levelplot(spc ~ .wavelength * z, log(barb), panel = panel.levelplot.points, cex = 0.3, col = "#00000000", col.regions = matlab.palette(20) ) }#>#> #>#> #> #>plotc(apply(barb[, , 42.9 ~ 43.2], 1, sum, na.rm = TRUE), spc ~ z, panel = panel.lines, ylab = expression(I[m / z == 43] / "a.u.") )