Spectra Plotting with ggplot2 Was Moved to hySpc.ggplot2 (DEPRECATED)
Source:R/DEPRECATED-ggplot2.R
hyperSpec-deprecated-ggplot2.RdThese ggplot2-related hyperSpec functions are deprecated
and they will be removed in the next release of the package.
Now functions from package hySpc.ggplot2
(link)
should be used as alternatives to plot hyperSpec objects with ggplot2.
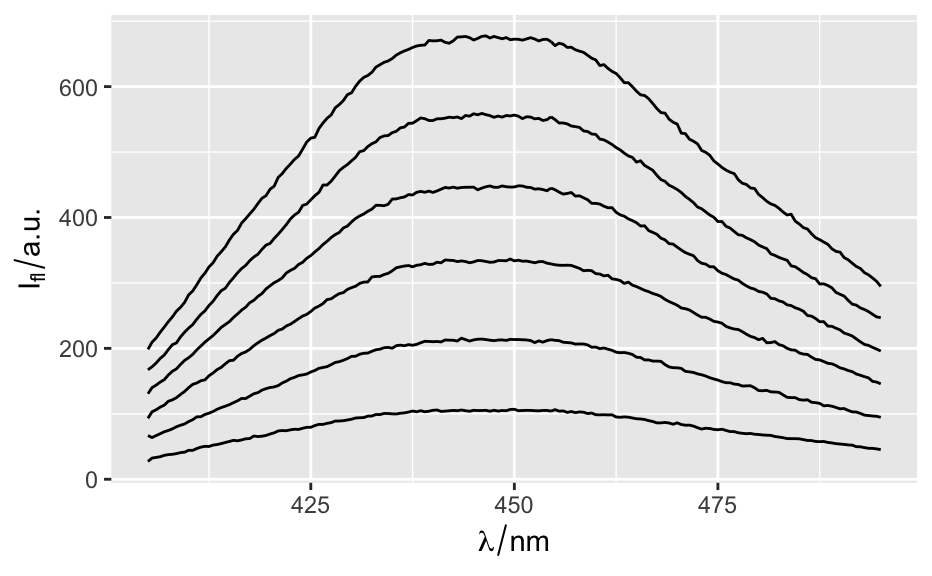
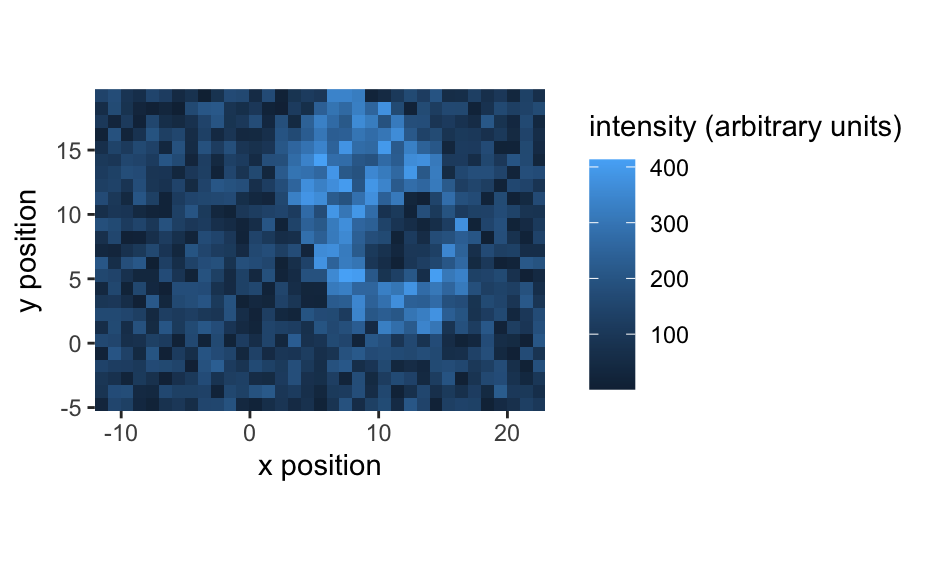
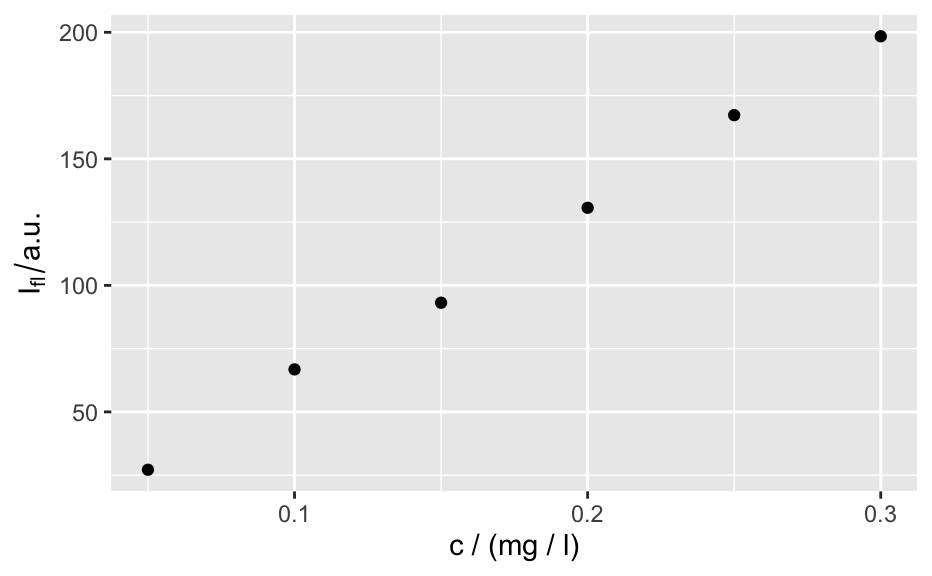
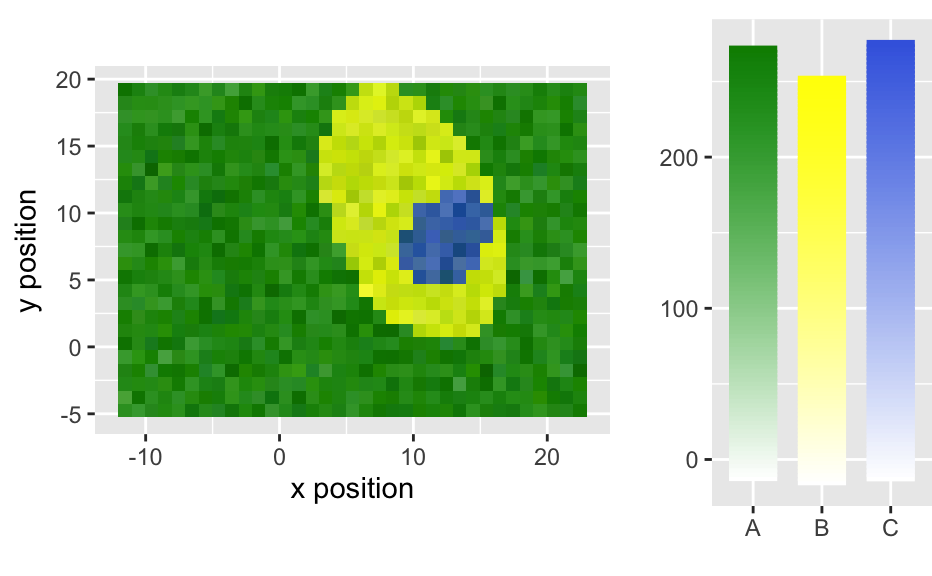
qplotspc( x, wl.range = TRUE, ..., mapping = aes_string(x = ".wavelength", y = "spc", group = ".rownames"), spc.nmax = hy.getOption("ggplot.spc.nmax"), map.lineonly = FALSE, debuglevel = hy.getOption("debuglevel") ) qplotmap( object, mapping = aes_string(x = "x", y = "y", fill = "spc"), ..., func = mean, func.args = list(), map.tileonly = FALSE ) qplotc( object, mapping = aes_string(x = "c", y = "spc"), ..., func = NULL, func.args = list(), map.pointonly = FALSE ) qplotmixmap(object, ...) legendright(p, l, legend.width = 8, legend.unit = "lines") qmixtile( object, purecol = stop("pure component colors needed."), mapping = aes_string(x = "x", y = "y", fill = "spc"), ..., map.tileonly = FALSE ) normalize.colrange(x, na.rm = TRUE, legend = FALSE, n = 100, ...) normalize.range(x, na.rm = TRUE, legend = FALSE, n = 100, ...) normalize.null(x, na.rm = TRUE, legend = FALSE, n = 100, ...) normalize.minmax(x, min = 0, max = 1, legend = FALSE, n = 100, ...) qmixlegend( x, purecol, dx = 0.33, ny = 100, labels = names(purecol), normalize = normalize.colrange, ... ) colmix.rgb( x, purecol, against = 1, sub = TRUE, normalize = normalize.colrange, ... )
Arguments
| x | matrix with component intensities in columns |
|---|---|
| wl.range | wavelength ranges to plot |
| ... |
|
| mapping | |
| spc.nmax | maximum number of spectra to plot |
| map.lineonly | if |
| debuglevel | if > 0, additional debug output is produced |
| object | matrix to be plotted with mixed colour channels |
| func | function to summarize the wavelengths, if |
| func.args | arguments to |
| map.tileonly | if |
| map.pointonly | if |
| p | plot object |
| l | legend object |
| legend.width, legend.unit | size of legend part |
| purecol | pure component colours, names determine legend labels |
| na.rm | see |
| legend | should a legend be produced instead of normalized values? |
| n | of colours to produce in legend |
| min | numeric with value corresponding to "lowest" colour for each column |
| max | numeric with value corresponding to "hightest" colour for each column |
| dx | width of label bar |
| ny | number of colours in legend |
| labels | component names |
| normalize | function to normalize the values. |
| against | value to mix against (for |
| sub | subtractive color mixing? |
Author
Claudia Beleites
Examples
qplotspc(flu)#> Warning: Function 'qplotspc' is deprecated. #> Use function 'qplotspc' from package 'hySpc.ggplot2' instead. #> https://r-hyperspec.github.io/hySpc.ggplot2qplotmap(faux_cell[, , 1200])#> Warning: Function 'qplotmap' is deprecated. #> Use function 'qplotmap' from package 'hySpc.ggplot2' instead. #> https://r-hyperspec.github.io/hySpc.ggplot2qplotc(flu)#> Warning: Function 'qplotc' is deprecated. #> Use function 'qplotc' from package 'hySpc.ggplot2' instead. #> https://r-hyperspec.github.io/hySpc.ggplot2#> Warning: Intensity at first wavelengh only is used.faux_cell <- faux_cell - spc.fit.poly.below(faux_cell) qplotmixmap(faux_cell [, , c(800, 1200, 1500)], purecol = c(A = "green4", B = "yellow", C = "royalblue") )#> Warning: Function 'qplotmixmap' is deprecated. #> Use function 'qplotmixmap' from package 'hySpc.ggplot2' instead. #> https://r-hyperspec.github.io/hySpc.ggplot2#> Warning: Removed 300 rows containing missing values (geom_point).