The plot method for hyperSpec objects is a switchyard to plotspc(),
plotmap(), and plotc().
# S4 method for hyperSpec,missing plot(x, y, ...) # S4 method for hyperSpec,character plot(x, y, ...)
Arguments
| x | the |
|---|---|
| y | selects what plot should be produced |
| ... | arguments passed to the respective plot function |
Details
It also supplies some convenient abbrevations for much used plots.
If y is missing, plot behaves like plot(x, y = "spc").
Supported values for y are:
- "spc"
calls
plotspc()to produce a spectra plot.- "spcmeansd"
plots mean spectrum +/- one standard deviation
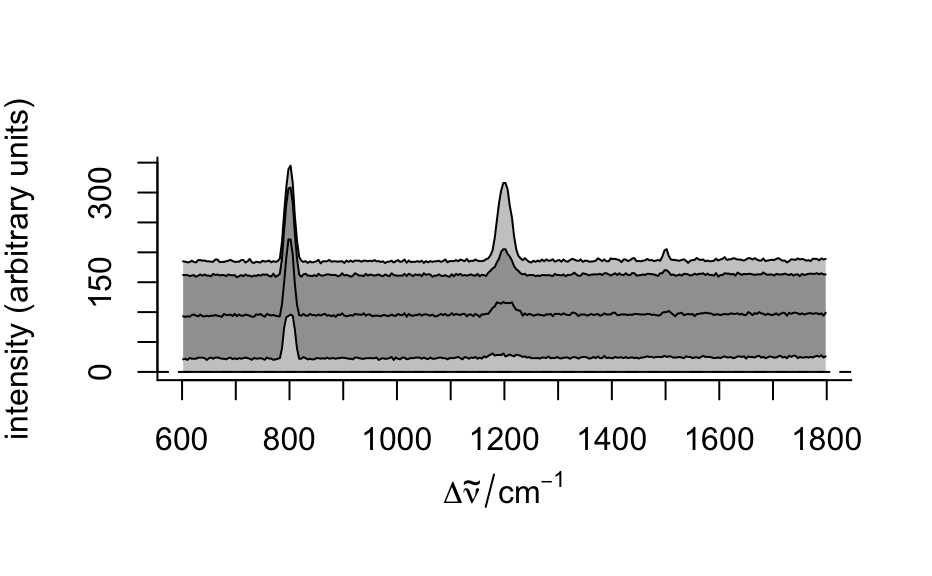
- "spcprctile"
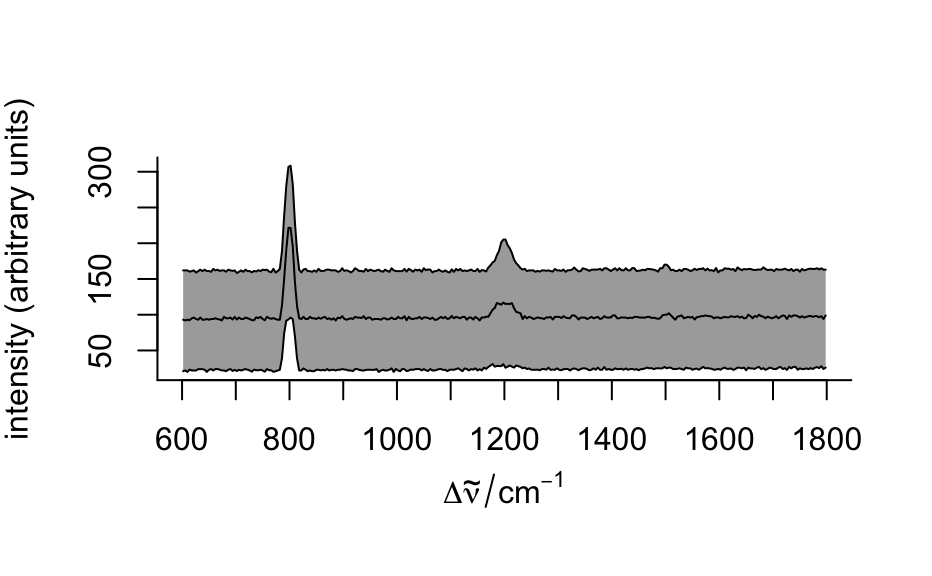
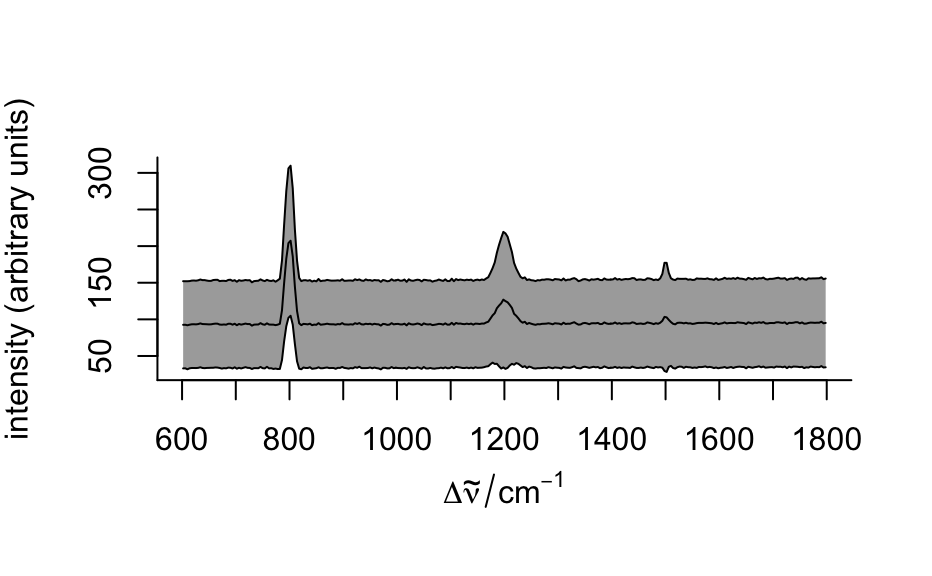
plots 16th, 50th, and 84th percentile spectre. If the distributions of the intensities at all wavelengths were normal, this would correspond to
"spcmeansd". However, this is frequently not the case. Then"spcprctile"gives a better impression of the spectral data set.- "spcprctl5"
like
"spcprctile", but additionally the 5th and 95th percentile spectra are plotted.- "map"
calls
plotmap()to produce a map plot.- "voronoi"
calls
plotvoronoi()to produce a Voronoi plot (tesselated plot, like "map" for hyperSpec objects with uneven/non-rectangular grid).- "mat"
calls
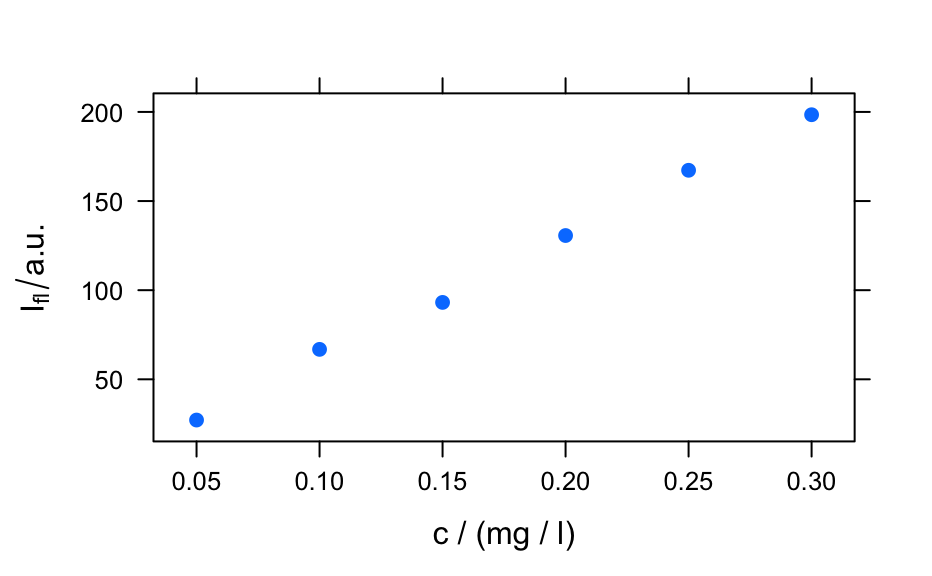
plotmat()to produce a plot of the spectra matrix (not to be confused withgraphics::matplot()).- "c"
calls
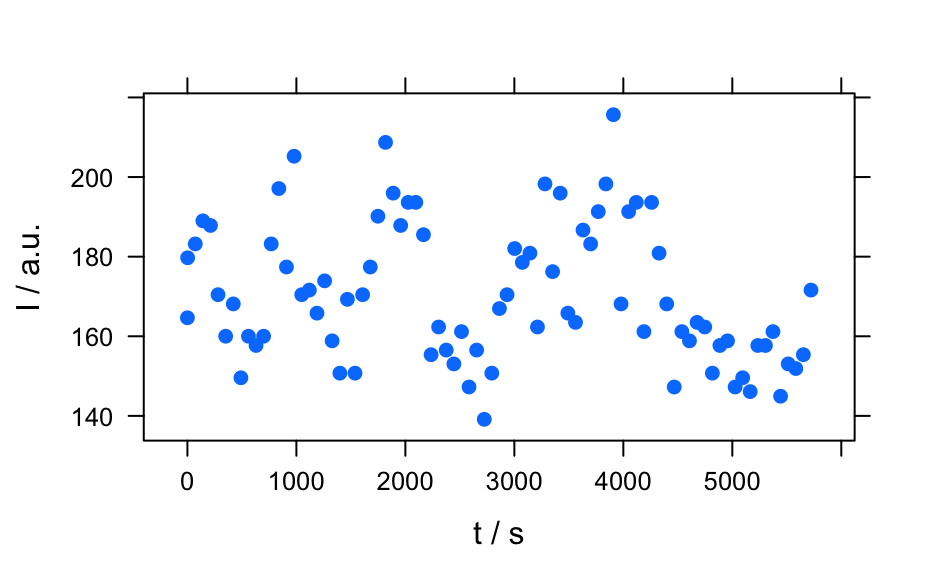
plotc()to produce a calibration (or time series, depth-profile, or the like).- "ts"
plots a time series: abbrevation for
plotc(x, use.c = "t").- "depth"
plots a depth profile: abbrevation for
plotc(x, use.c = "z").
See also
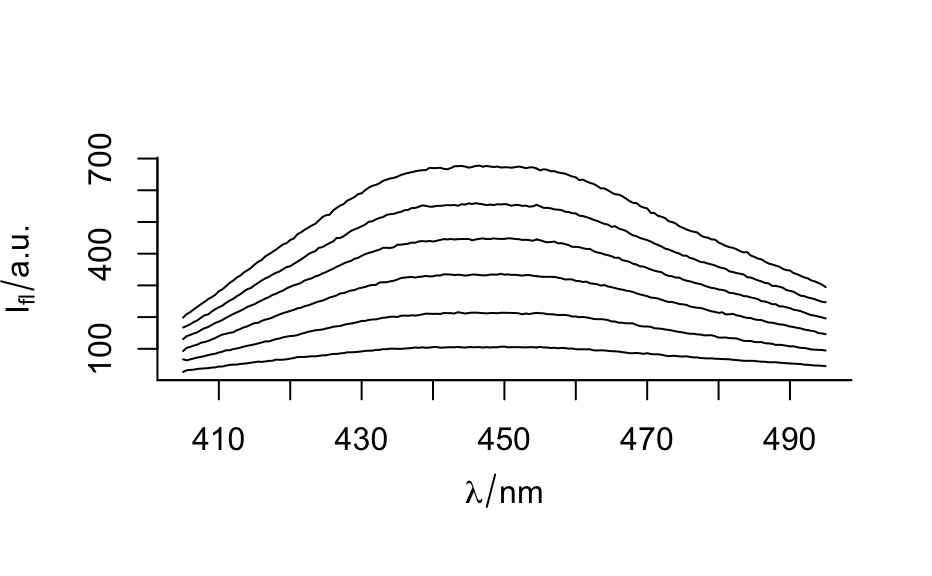
plotspc() for spectra plots (intensity over wavelength),
plotmap() for plotting maps, i.e. color coded summary value on two
(usually spatial) dimensions.
plotc()
Author
C. Beleites
Examples
plot(flu)#> Warning: Intensity at first wavelengh only is used.#> Warning: Intensity at first wavelengh only is used.spc <- apply(faux_cell, 2, quantile, probs = 0.05) spc <- sweep(faux_cell, 2, spc, "-") plot(spc, "spcprctl5")### use plotspc as default plot function