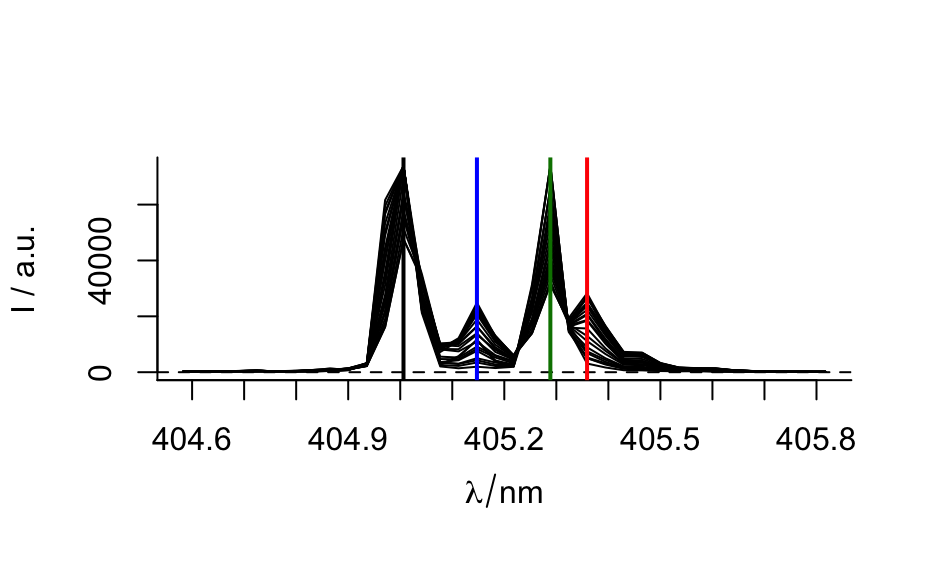Calibration- and timeseries plots, depth-profiles and the like
plotc plots intensities of a hyperSpec object over another
dimension such as concentration, time, or a spatial coordinate.
plotc( object, model = spc ~ c, groups = NULL, func = NULL, func.args = list(), ... )
Arguments
| object | the |
|---|---|
| model | the lattice model specifying the plot |
| groups | grouping variable, e.g. |
| func | function to compute a summary value from the spectra to be plotted instead of single intensities |
| func.args | further arguments to |
| ... | further arguments to |
Details
If func is not NULL, the summary characteristic is calculated
first by applying func with the respective arguments (in
func.args) to each of the spectra. If func returns more than
one value (for each spectrum), the different values end up as different
wavelengths.
If the wavelength is not used in the model specification nor in
groups, nor for specifying subsets, and neither is
func given, then only the first wavelength's intensities are plotted
and a warning is issued.
The special column names .rownames and .wavelength may be used.
The actual plotting is done by lattice::xyplot().
See also
Author
C. Beleites
Examples
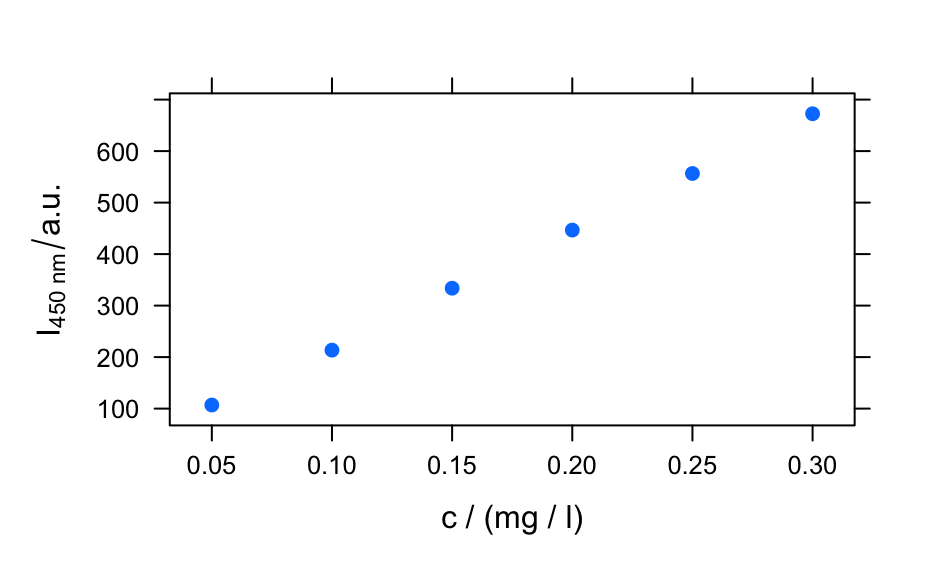
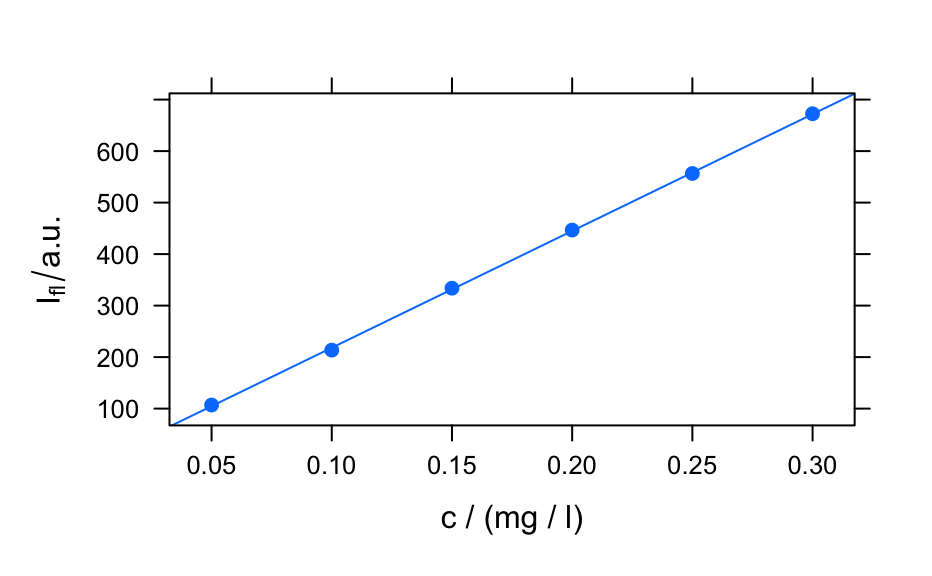
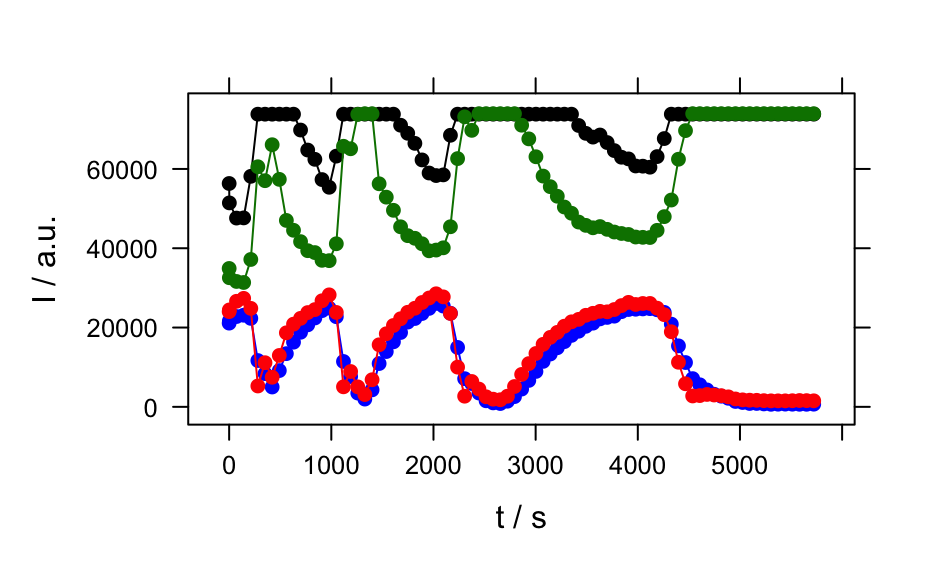
## example 1: calibration of fluorescence plotc(flu) ## gives a warning#> Warning: Intensity at first wavelengh only is used.plotc(flu, func = mean)plotc(flu, func = range, groups = .wavelength)#> #> Call: #> lm(formula = spc ~ c, data = flu[, , 450]$.) #> #> Residuals: #> 1 2 3 4 5 6 #> 2.1918 -4.6846 2.1696 1.6005 -1.9306 0.6533 #> #> Coefficients: #> Estimate Std. Error t value Pr(>|t|) #> (Intercept) -8.666 2.876 -3.013 0.0394 * #> c 2268.482 14.769 153.596 1.08e-08 *** #> --- #> Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1 #> #> Residual standard error: 3.089 on 4 degrees of freedom #> Multiple R-squared: 0.9998, Adjusted R-squared: 0.9998 #> F-statistic: 2.359e+04 on 1 and 4 DF, p-value: 1.078e-08 #>conc <- list(c = seq(from = 0.04, to = 0.31, by = 0.01)) ci <- predict(calibration, newdata = conc, interval = "confidence", level = 0.999) panel.ci <- function(x, y, ..., conc, ci.lwr, ci.upr, ci.col = "#606060") { panel.xyplot(x, y, ...) panel.lmline(x, y, ...) panel.lines(conc, ci.lwr, col = ci.col) panel.lines(conc, ci.upr, col = ci.col) } plotc(flu[, , 450], panel = panel.ci, conc = conc$c, ci.lwr = ci[, 2], ci.upr = ci[, 3] )## example 2: time-trace of laser emission modes cols <- c("black", "blue", "#008000", "red") wl <- i2wl(laser, c(13, 17, 21, 23)) plotspc(laser, axis.args = list(x = list(at = seq(404.5, 405.8, .1))))plotc(laser[, , wl], spc ~ t, groups = .wavelength, type = "b", col = cols )